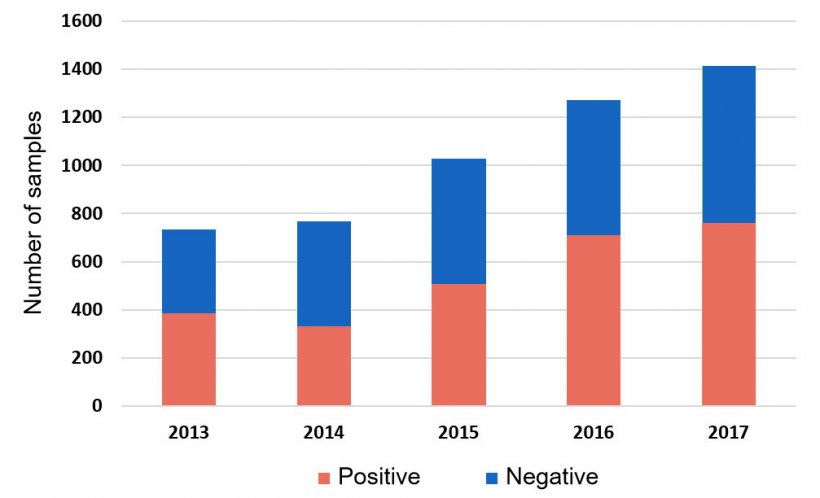
Haemophilus parasuis (HPS) continues to challenge swine producers causing fibrinous polyserositis, meningitis and arthritis, syndrome commonly known as Glässer’s Disease. It is considered one of the most prevalent bacterial infections during the nursery stage, and regular medication to control this disease is an emerging concern. In fact, more than 50% of all systemic samples tested by HPS PCR at the Iowa State University Veterinary Diagnostic Laboratory (ISU VDL) are positive (Figure 1).

In terms of vaccination, whole-cell killed products have shown, in general, to be protective only against strains of the same serotype. HPS is a highly diverse pathogen, and 15 serotypes have been described by agar gel precipitation test (AGPT) and indirect hemagglutination (IHA) (Turni and Balckall, 2005). These serotyping methods are based on reactions between surface antigens, such as capsular polysaccharides, and antisera. Even though traditional serotyping is the most commonly used method to characterize HPS, a considerable number of isolates are untypeable.
Further progress in molecular methods have led to the development of a multiplex serotyping PCR (Howell et al., 2015; Lacouture et al., 2017). This PCR reaction uses specific primers based on serotype-specific variations on the capsule loci, capable of discriminating between 14 of 15 HPS serotypes. No capsular polysaccharide gene could be confirmed differentiate between serotypes 5 and 12. In 2017, Jia and collegues proposed a new HPS serotype-specific PCR scheme, particularly containing primers that differentiate serotype 5 from serotype 12. However, the primer for serotype 12 is based on a hypothetical gene, and further investigations are warrented.
Comparison between serotyping PCR and IHA showed that both techniques had the same results for 88% of the HPS isolates tested. The differences observed could be explained by complications with the IHA method. The most striking difference is that the number of untypable strains (strains not being able to be serotyped by the use of antisera) was significantly reduced Howell et al., 2015; Lacouture et al., 2017). Serotyping PCR has been optimized and it is now being offered by the ISU VDL.
When a subset of 216 HPS strains isolated from porcine samples submitted to ISU VDL were analyzed, only three isolates (0.5%) were untypeable by serotyping PCR, which confirms an important reduction when compared to the conventional methods (15-41%). Serotypes 4 (24%), 7 (26.7%), 1 (14.8%), 2 (13.4%), 5/12 (13%) and 13 (10.2%) were the most commonly identified, followed by serotypes 14 (5.6%) and 6 (0.9%) (Figure 2). These serotypes have been also commonly identified from the field in the US, Canada, Europe and China.
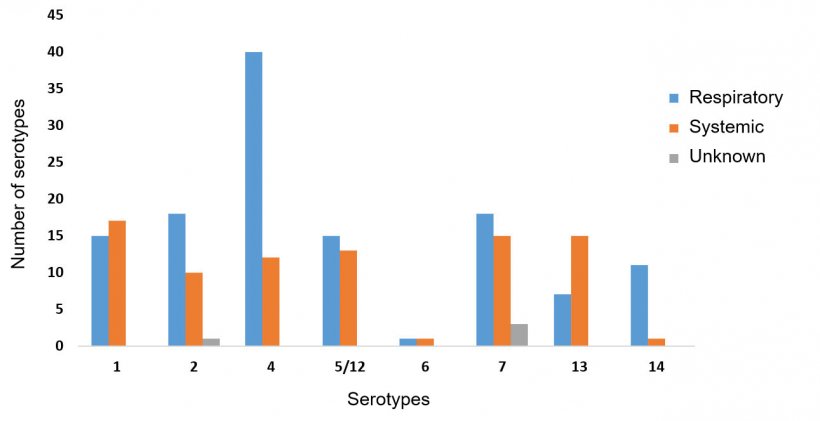
Most HPS isolates from this collection were obtained from diseased pigs affected by pneumonia and polyserositis. The site of HPS isolation was primarily pulmonary (Figure 2), followed by systemic sites such as pericardium (21%), pleura (7%), joint (5%), meninge (3.7%), peritoneum (1.4%) and fibrin (0.5%). This fact strongly suggests that those isolates were virulent for being able to resist serum killing and phagocytosis, both recognized virulence mechanisms in HPS (Cerda-Cuellar & Aragon, 2008; Olvera et al., 2009). Even serotypes 6 and 7, previously classified as non-virulent by the Kielstein-Rapp-Grabrielson scheme (Kielstein & Rapp-Gabrielson, 1992), were isolated from systemic sites. HPS can be isolated from lungs of healthy pigs, and coinfections are common. Therefore, the presence of other potentially virulent strains infecting the same animal should be considered.
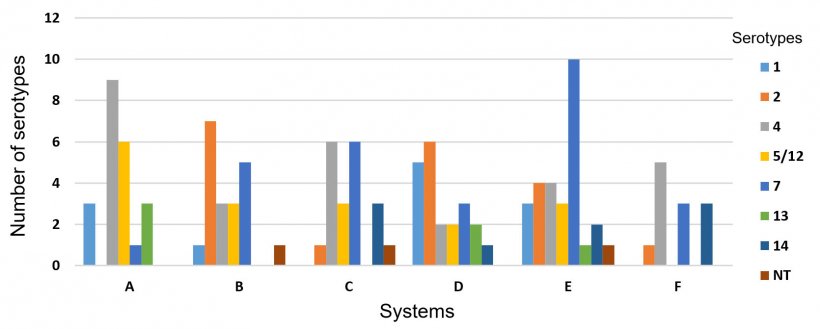
One hundred and twenty three HPS isolates recovered from six different swine systems in the USA were analyzed separately to evaluate the frequency of serotypes within systems. A high variability of serotypes was observed in those systems, particularly because each system is composed of several different farms. However, in most cases only one serotype prevailed within a flow. Also, we found that serotypes 4 (23.5%, 29/123) and 7 (22.7%, 28/123) were present in all systems (Figure 3).
Control of HPS requires a multifaceted approach. The first step is to characterize the isolates found in diseased pigs. Monitoring the presence of HPS serotypes within herds is essential for disease control. Serotyping PCR and VtaA PCR can be useful tools particularly with regard to selection of virulent serotypes for inclusion in autogenous vaccines, to improve pig flow management by commingling sources with the same serotype, and to prevent introduction of pigs with potentially virulent HPS strains into naïve herds.



