Vaccination against porcine circovirus type 2 (PCV2) successfully protects against porcine circovirus associated disease (PCVAD). PCV2 vaccines were introduced into the United States in 2006 following an increase in PCVAD cases and were rapidly adopted throughout the swine industry to control PCVAD outbreaks. Implementation of intensive vaccination in the US since 2006 has induced high levels of anti-PCV2 antibodies and reduced viral loads, but has not completely eliminate virus from farms (Dvorak et al, 2016). Continued vaccination may lead to PCV2 elimination in herds, but virus continues to be observed in one third to one half of the sites across the US in spite of regular vaccination (Dvorak et al, 2016).
All major PCV2 vaccines today are based on the PCV2a genotype, but have been shown to protect against disease caused by other genotypes (Fort et al, 2008; Opriessnig et al, 2017). A shift in the major genotype observed over the years from PCV2a to PCV2b, and now to PCV2d, was thought to be linked to the use of PCV2a genotype vaccines. However, outbreaks of PCVAD seemingly associated with newly emerging genotypes were controlled after rigorously re-instituting the same or a modified vaccination program. This suggests that there was not a failure of immunity to cross-protect, but that the disease outbreak was due to inadequate vaccine compliance, such as improper storage, improper application, or similar issues at the farm. To examine the shift in genotypes more closely, we obtained the PCV2 sequence database from the University of Minnesota Veterinary Diagnostic Laboratory (UMN-VDL) for examination of PCV2 epidemiological changes following widespread use of vaccine.

Results
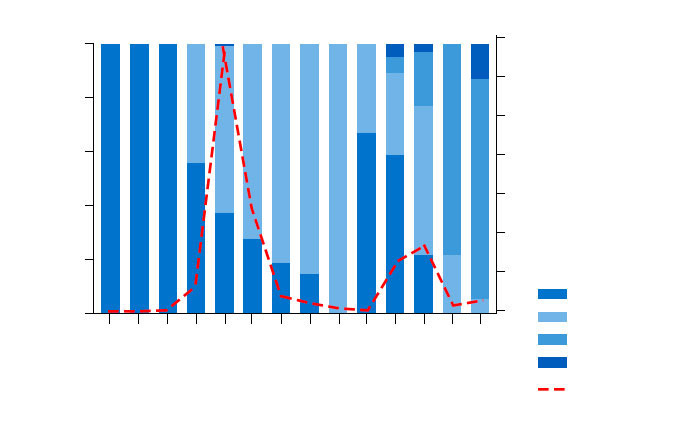
PCV2 ORF2 sequences (Genbank ID KT867794-KT868522, n=729) from field isolates submitted to the UMN-VDL from 2002 – 2015 were verified for completeness, accuracy, and associated metadata (Davies et al. 2016). The majority of sequences were obtained in 2006 (47%) and 2007 (18%) and another large group of sequences were obtained from 2012-2013 (Figure 1). PCV2a was the only genotype present in the examined samples until 2005 when PCV2b began to be observed (Figure 1). The major genotype seen from 2006 – 2010 was PCV2b, but in 2011 and 2012, a resurgence of PCV2a was observed (Figure 1). PCV2d sequences were first identified in 2012 and by 2014 had become the major genotype (Figure 1). PCV2e isolates were first observed in 2006 and then again in 2012, but were present at low prevalence (Figure 1).
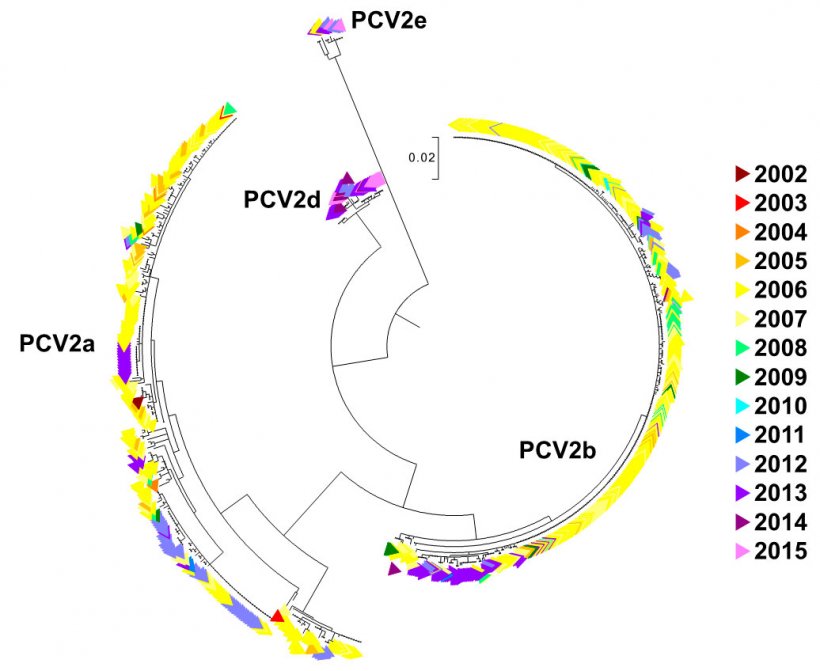
In order to visually illustrate the distribution of genotypes over time, a maximum likelihood tree was created using MEGA 7.0.21 and was color coded according to year (Figure 2, n=729). Interestingly, both PCV2a andPCV2b genotypes have a continuous presence throughout the time period examined, with the diversity of sequences within each genotype decreasing. A few clusters of identical or nearly identical sequences were present, most notably for PCV2b in the years 2006-2007, and then appeared to die out (Figure 2). PCV2d and PCV2e genotypes were more recently observed (2012 – 2015), although PCV2e was also present in 2006 (Figure 2).
Conclusions
PCV2 vaccines were first introduced into the United States swine herd in 2006 and are based on the PCV2a genotype. Vaccination clearly is effective in eliminating PCVAD and in substantially reducing viremia levels. However, vaccination may not eliminate PCV2 from herds and may result in a higher prevalence of non-PCV2a genotypes. Examination of >700 well-curated PCV2 sequences from the UMN-VDL database shows that an increase in the prevalence of the PCV2b genotype virus began in 2005, before vaccines were available. The resurgence of PCV2a genotype viruses in 2011 indicates that PCV2a was present in farms across the U.S. in spite of vaccination. Thus, the decrease in PCV2a and PCV2b genotypes over time and the increase in PCV2d in 2012 may be due more to other, unknown, factors than to immunological selection resulting from PCV2a-based vaccination.




