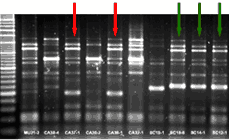
 |
| ERIC-PCR of nasal strains of Haemophilus parasuis obtained from 3 different farms. The isolated bacterium with the same pattern of bands are from the same strain (indicated with arrows of the same colour). |
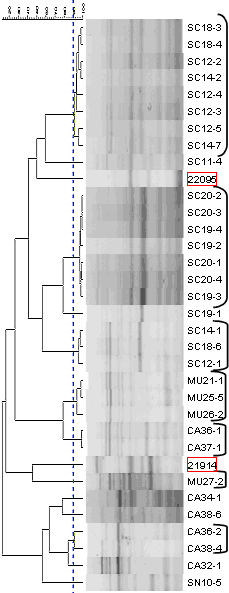
 |
| The patterns of ERIC-PCR can be analysed with special programmes that allow the comparison of different isolations. Diverse nasal strains are compared with each other and with isolated strains of lesions (in red boxes). The patterns are considered to be equal up to a level of 90º of identity (blue dotted line). |
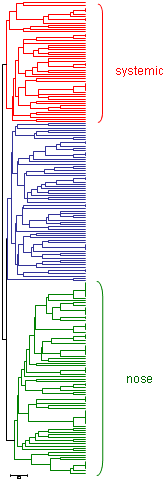 |
| Classification of 211 strains of Haemophilus parasuis using MLST. There is one group that is associated to isolated strains of systemic lesions (in red), another group associated with isolated strains of nose of healthy animals (in green) and a last intermediate group (in blue). |
Therefore, it is important to be able to identify the distinct circulating strains and to differentiate between the strains that are merely colonisers and those that have the capacity to produce disease. With this aim various methods of classification of strains or “typing” of H. parasuis have been developed.
The typing methods can be useful in local or global epidemiologic studies. Local studies concentrate on analysing the strains that are present on one farm or on related farms, determining the introduction of new strains or checking to see if a treatment has eliminated the originating strain of an outbreak. Whereas, the global studies analyse the relationship of certain strains with others that have been isolated in distinct times and places.
Classically, the strains of H. parasuis have been classified by serotyping. In 1992, Kielstein and Rapp-Gabrielson established 15 serotypes in H. parasuis and determined the virulence of the reference strains of the distinct serotypes by experimental infection. According to this classification, the most virulent serotypes are 1, 5, 10, 12, 13 and 14. In studies carried out in different countries, including Spain, it was observed that the most prevalent serotypes are 5 and 4, with 4 being of moderate virulence. Although serotyping has the advantage of proportioning information on the antigenic reaction of the strains, its biggest problem is that there are a great number of strains that are not typeable (between 15 and 41%).
In order to avoid this problem methods of genotyping began to be developed which analyse genetic material present in all the strains. The biggest disadvantage of genotyping is the lack of direct information so that the classifications obtained have to be complemented with other data, such as the clinical origin of the strain. There are various genotyping methods that are based both on the comparison of the band profiles and on the comparison of sequences.
One example of the method of band comparison used in the classification of H. parasuis strains is ERIC-PCR (“enterobacterial repetitive intergenic consensus” PCR). This technique is useful in local studies of H. parasuis strains, but its reproducibility is not good and the results from different laboratories cannot be compared. In order to improve the genotyping, sequencing methods have been developed such as “multilocus sequence typing” or MLST. With MLST, the H. parasuis strains were classified into various groups which were related to the potential virulence of the strains. One group included strains associated to systematic disease and virulent reference strains, while another group included nasal strains of healthy animals and non-virulent reference strains.
Genotyping methods have a great potential for the performance of epidemiological studies of H. parasuis, but more effort is required to relate the genomic classification and the real virulence of the strains.



