PCR samples and future perspectives of Third Generation Sequencing as a diagnostic tool for SMEDI complexes and abortions in sows
Background and Objective
Abortion and SMEDI (Stillbirth, Mumification, Embryonic Death, Infertility) represent a significant challenge in pig farming worldwide with root cause analysis on the farms always including non-infectious as well as infectious agents. Infection diagnostic approaches encompass both facultative pathogenic and obligate pathogenic agents. The wide range of possible infectious causes combined with the challenging diagnostic timing - especially in SMEDI cases - leads to significant uncertainty regarding reliable result interpretation. This work aims to compare classical PCR methods through a retrospective analysis of samples collected over the past two years (2022 to 2023) while also exploring early results from Third Generation Sequencing. The focus is to assess whether sequencing technology could offer valuable insights and provide a potential advantage in future diagnostic approaches for abortion and SMEDI cases in swine production.

Material and Methods
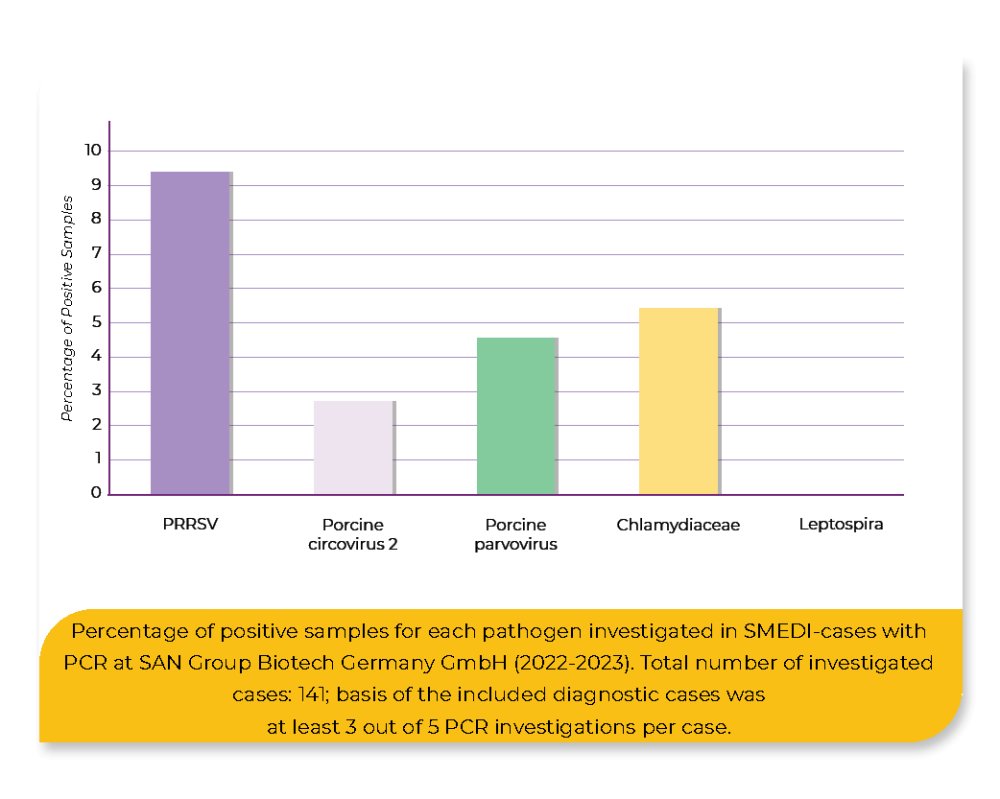
A retrospective analysis was conducted by analyzing 141 cases of submitted fetal tissue and/or placenta collected from routine diagnostics in 2022 and 2023 at SAN Group Biotech Germany GmbH. All samples had previously been screened with real-time PCR for at least three of the following pathogens: Porcine Parvovirus (PPV; 114 investigations; KYLT PPV), Porcine Reproductive and Respiratory Syndrome Virus (PRRSV; 141 investigations; LSI VetMAXTM PRRSV EU/NA), Porcine Circovirus 2 (PCV-2; 127 investigations; KYLT PCV-2), Chlamydia spp. (93 investigations; KYLT Chlamydiaceae Screening) and Leptospira spp. (118 investigations; KYLT pathogenic Leptospira).
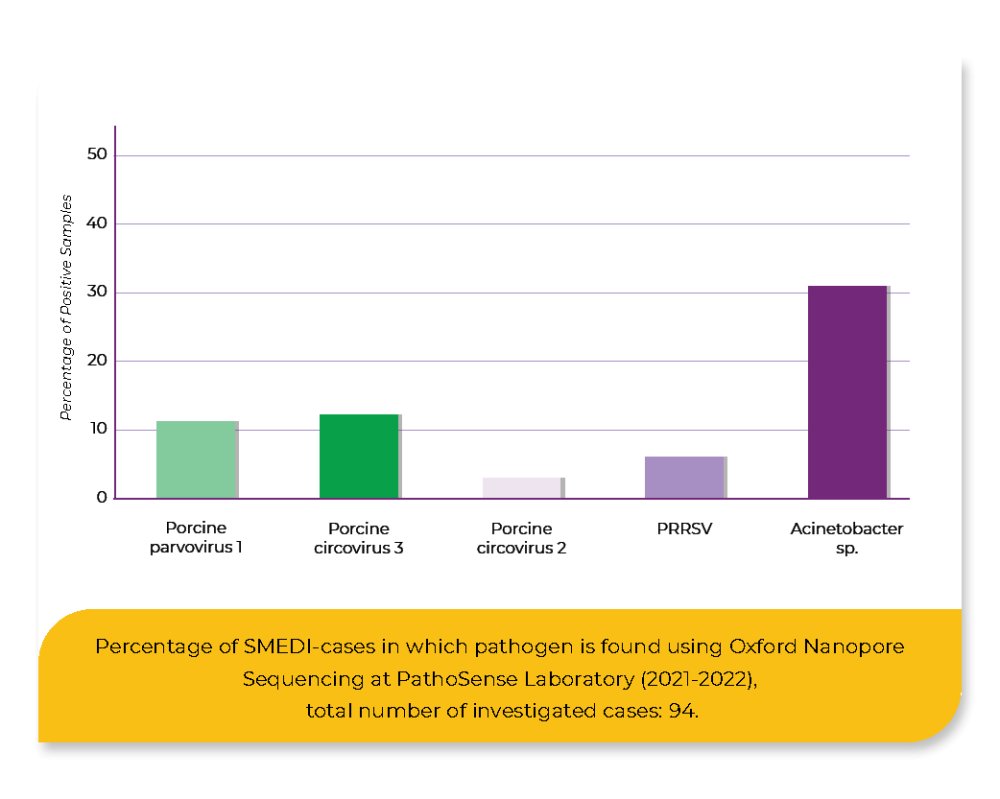
Another 94 samples collected in 2021 and 2022, mostly SMEDI cases, were analyzed by PathoSense using a novel viral and bacterial metagenomics workflow using nanopore sequencing.
Results
By using the real-time PCR, 18.4% positive samples could be detected, showing that PRRSV was the most frequently detected pathogen (9.2%), followed by Chlamydia spp. (5.4%), PPV (4.4%) and PCV-2 (2.2%). Leptospira spp. was not detected. Using nanopore sequencing, 61.7% of the samples were tested positive, comprising PPV1 (11.7%), PCV-3 (12.8%), PCV-2 (3.2%), PRRSV (6.4%) and Acinetobacter sp. (33%). The remaining 38.3% of the samples were tested negative.
Discussion and Conclusion
For many years, the routine diagnostic approach for investigating cases of abortion and SMEDI in swine has relied on PCR screening for the most commonly known pathogens, such as PRRSV, PPV, PCV-2, Leptospira, and Chlamydia. However, when comparing this traditional method with Third Generation Sequencing, a different spectrum of pathogens has been identified. Even though different samples were investigated for both approaches, the result raises important questions about whether conventional diagnostics may have overlooked additional, potentially relevant infectious agents. While sequencing has clearly expanded the range of detectable pathogens, further research is needed to determine the clinical significance and pathogenic role of such. With this comparison we aim to highlight the potential of Third Generation Sequencing as a future tool for more comprehensive pathogen identification in reproductive disorders research, ultimately offering insights into more effective prevention and treatment strategies for reproductive losses in swine herds. In addition, possibilities can be shown and discussed as to how both molecular biological methods can be used effectively together.
Let’s talk solutions and connect with our corresponding author:
juhle-marijke.buch@san-group.com
Credits:
J. Buch1, I. Spiekermeier1, S. Coppens2, S. Theuns2
1 SAN Group Biotech Germany GmbH, Hoeltinghausen, Germany
2 PathoSense Laboratory, Ghent, Belgium
SAN Group Biotech Germany GmbH
Mühlenstraße 13 I 49685 Höltinghausen GERMANY
+49 4473 9438 0 I office-de@san-group.com
Contact:
Contact us using the following form.

